L-Asparaginase Sequences to Treat Leukemia
Drug Discovery with Ancestral Sequence Reconstruction
Leukemia: It’s one of the most terrifying words a parent can hear, yet acute lymphoblastic leukemia (ALL) is the most common childhood cancer, accounting for about 25% of all childhood cancers in the United States. ALL is an aggressive cancer, but there are viable treatment options currently available. However, 30% of the patients who receive the current therapeutic (L-asparaginase) experience severe toxicity as L-asparaginase is essentially derived from bacteria. The patients can also experience hypersensitivity, pancreatic and liver damage, and adverse immunological responses, which happen when the immune system reacts to the therapeutic as though it were an allergen.
Using Genomic Ancestry to Chart a Path Forward
Asparaginase is an enzyme that breaks down asparagine, an amino acid necessary for protein synthesis in leukemic cells. In short, asparaginase increases the probability that the cancerous cells will die. It has been the first-choice therapeutic for ALL since the 1970s. However, there was a mass shortage of the drug in 2016, which, in combination with the list of adverse side effects, inspired Emory researchers Sunil Raikar, MD, and Christopher Doering, PhD, to find novel forms of asparaginase.

Sunil Raikar, MD
With their combined expertise in genetics, hematology, and oncology, Raikar and Doering sought to find a form of asparaginase that would be more compatible with human biochemistry. According to Raikar, the 2016 shortage made them ask, “Can we make a drug that is more humanized, or closer to human asparaginase?”
The first piece of the puzzle came from Eric Gaucher, a molecular evolutionist at Georgia Tech. He developed a computational method for backtracking the evolution of proteins to find similarities between species. The ancestral sequence reconstruction process starts with a protein that has similar functionality across different species (referred to as an ortholog). This protein is used to build a phylogenetic tree, which connects the protein and its interspecies variants to a common ancestor. Evolutionists can then use this phylogenetic tree to predict an estimated protein sequence at a given time in history.

Christopher Doering, PhD
Doering and Raikar considered how this platform could be applied to making therapeutics by finding new functional proteins. Designer proteins are expensive (an average of $5,000 each), and there’s no certainty that they will even have the properties for which they were designed. However, Doering and Raikar found that the protein sequences predicted by the platform’s algorithm were plausible and functional.
“We thought maybe we could use [the platform] as a discovery tool to try to find new [protein] properties that we didn’t know about already,” stated Doering.
Armed with a new method of drug discovery and a mission to find a more humanized form of asparaginase, the researchers came to the second piece of the puzzle: guinea pigs.
Finding Alternative Asparaginase in Mammalian Protein
Though humans can produce asparaginase, it’s usually not in effective quantities, which is why asparaginase therapeutics are needed to treat certain cancers. Even though they’re FDA-approved, the current asparaginase therapeutics cause severe side effects for many patients.
Raikar and Doering had the difficult task of finding a mammalian asparaginase enzyme sequence that displayed the desired properties (i.e. compatible with human biology, effective activity, functional, etc.).
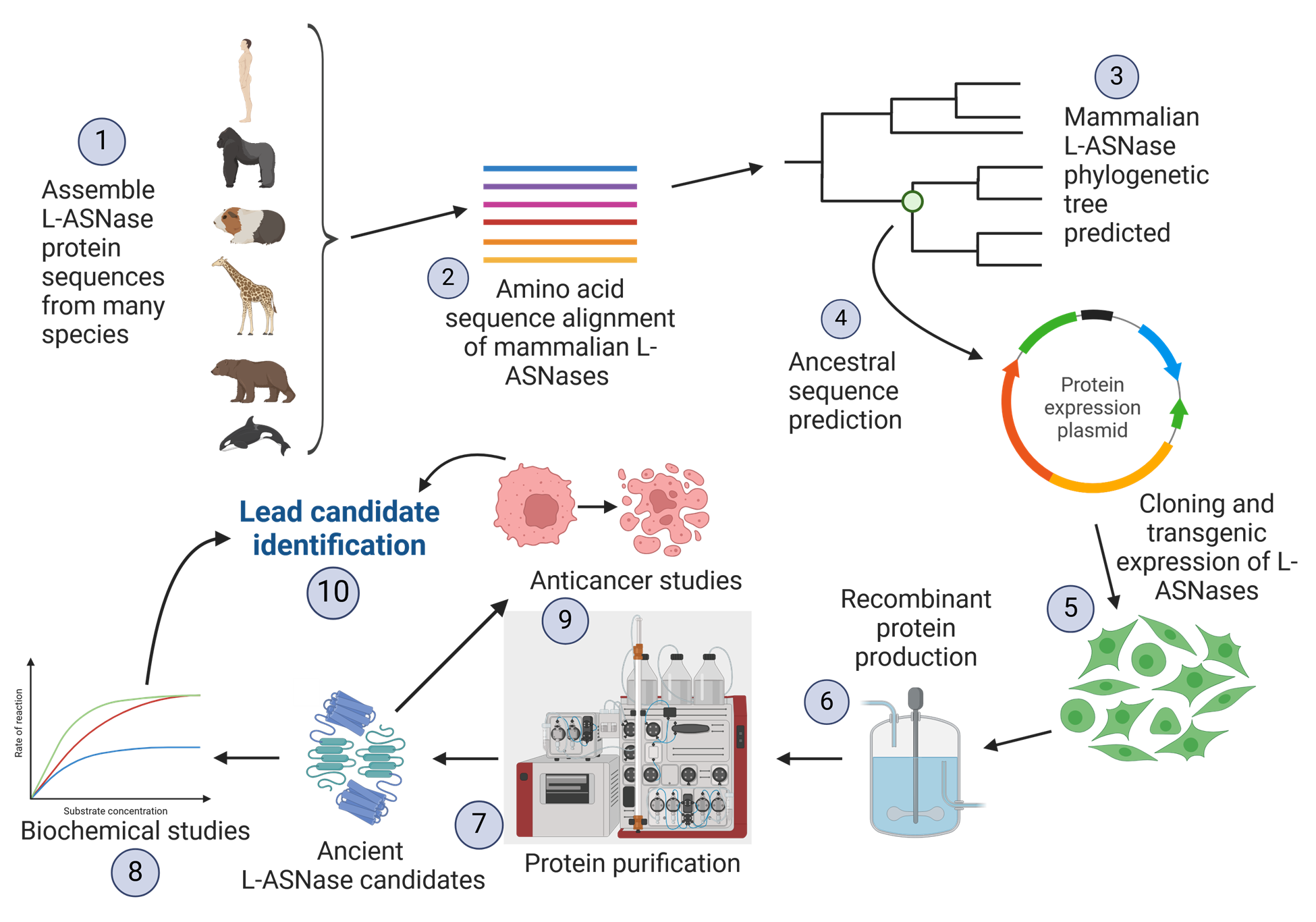
The process of finding alternative asparaginase in mammalian protein.
Illustration courtesy of the inventors
According to Doering, “If you look at proteins derived from the blood of different animals and compare that to the proteins from the blood of humans, you find that they have the same overall activity, but they have different properties in how well they do that activity.”
As the researchers would discover, guinea pigs have a highly functional asparaginase sequence that is about 70% closer to the human sequence than the bacterial sequence used in current therapeutics. The discovery platform found that there are about ten evolutionary protein sequences between guinea pigs and humans that show promising activity against cancer cells.
Taking the Next Steps
The USPTO granted Doering and Raikar’s patent in December 2024. With research in the preclinical stages, their goal now is to narrow their ten potential proteins to one candidate for further development. Regarding commercializing their invention, they have considered forming their own start-up and have received interest from investors.
The importance of their work is not just in its translational value. Before their work with asparaginase, there were five drug discovery techniques for protein therapies. Now, there are six, and their method is efficient and cost-effective. Their work has implications for treating ALL, and it has also changed the way researchers can discover and explore new drugs and therapies.
— Jenna Woods
Tech ID: 22185
Read the technology brief.